The goals of this tool are to automate the process of performing target direct genome mining1,2,3, search for potential novel bioactive compound targets, and prioritize putative secondary metabolite gene clusters. The following initial automated steps, and associated genome mining tools, that are used to realize these goals are:
- Predict Secondary Metabolite gene clusters: Antibiotics & Secondary Metabolite Analysis SHell (antiSMASH)
- Identify known antibiotic targets & Domains of Unknown Function: Pfam models of essential genes known to be targeted from published work & DUF domains
- Identify known resistance factors: ResFams, manually curated models that include proteins from The Comprehensive Antibiotic Resistance Database (CARD), The LACtamase Engineering Database (LACED), and The Jacobi and Bush Collection
- Identify essential genes: FunARTS comparative pipeline + BUSCO v5.4.3 and OrthoDB v10 Equivologs
Upon completion, the FunARTS workflow uses these results to:
- Proximity check: Cross reference locations with Secondary Metabolite gene clusters
- Uncommon duplication check: Highlight potential repurposed primary metabolism genes
- Visualize results: Provide an interactive format for rapid manual confirmation
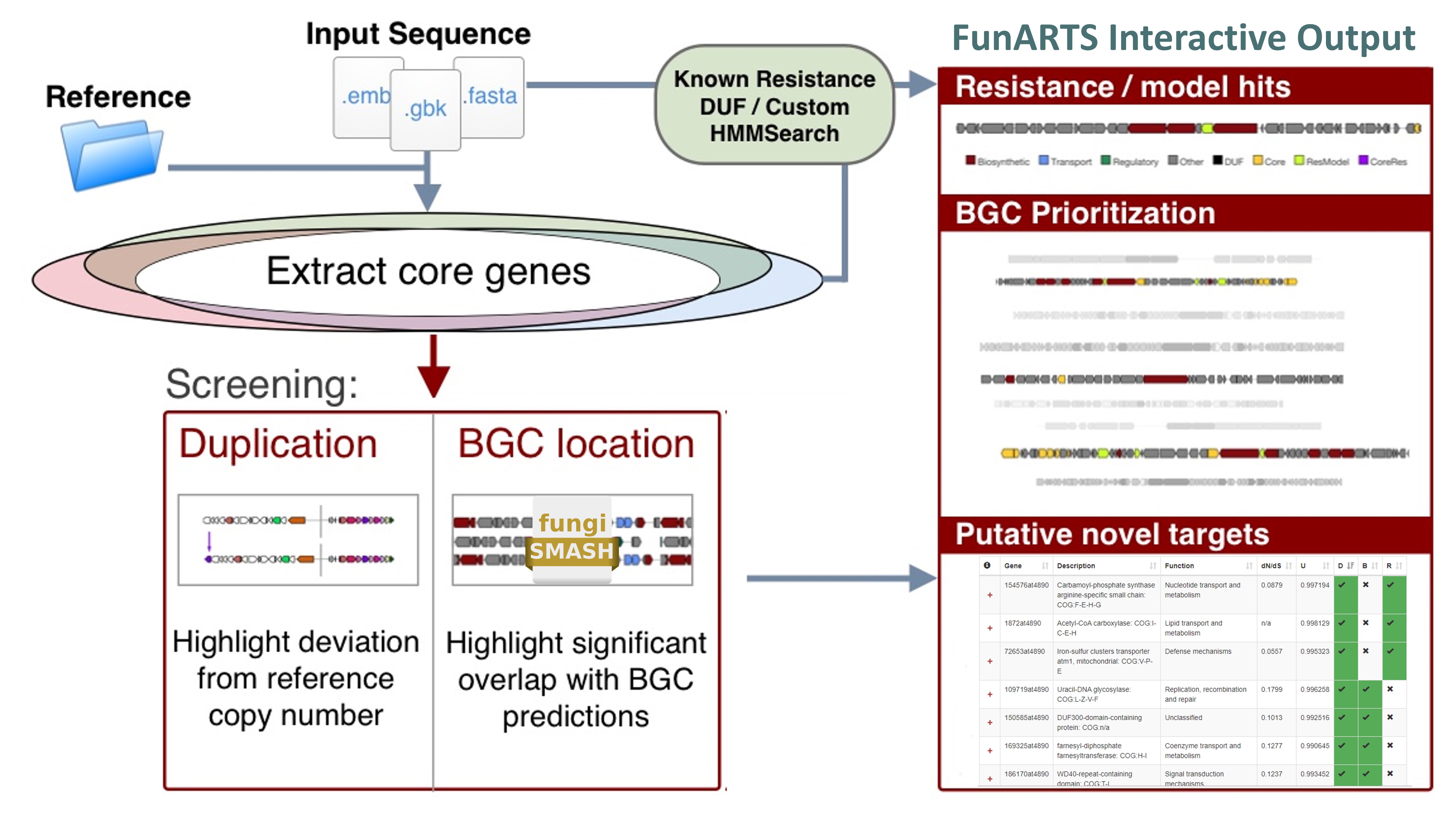
Potential novel targets are highlighted by exploring the space of essential genes in a query genome and filtering them by criteria associated with antibiotic resistance:
- Co-expression with secondary metabolite gene cluster: Self-resistance method to avoid suicide during production
- Duplication: To maintain non-resistant, likely higher fitness, version of gene
Reference workflow:
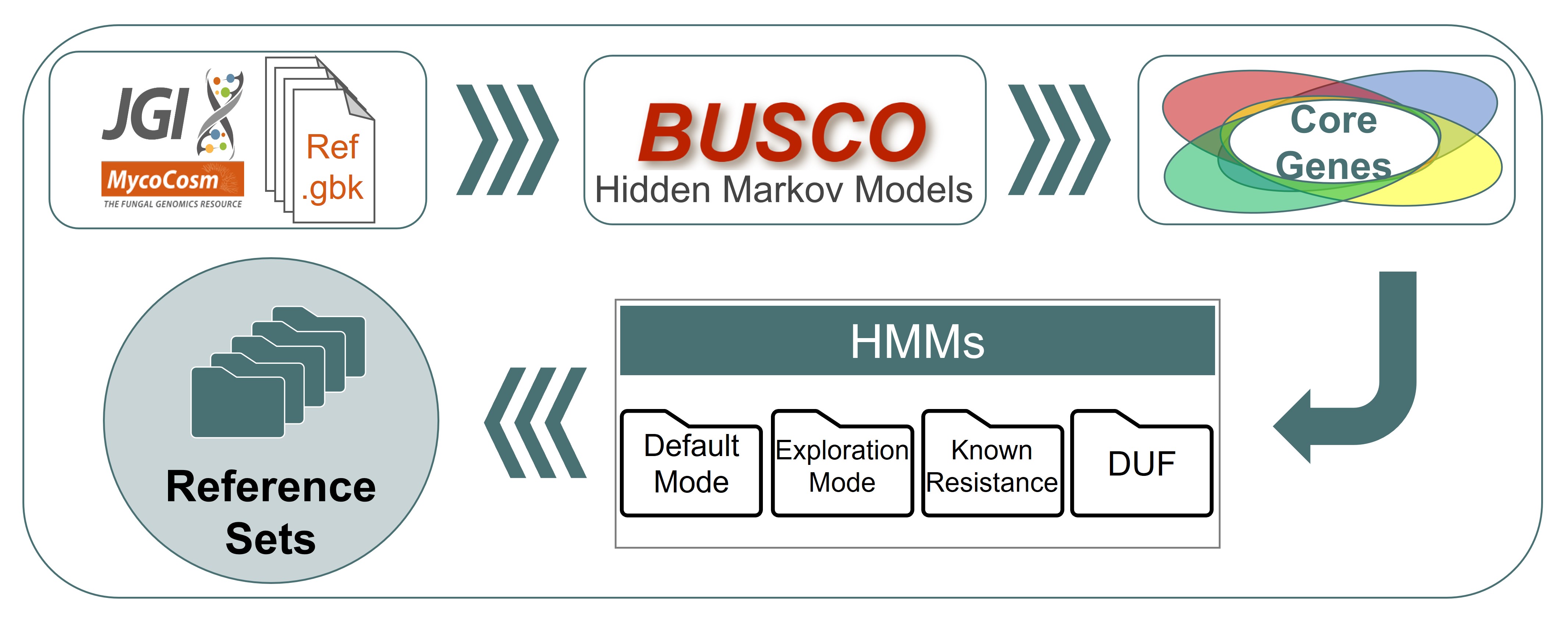
- Reference genomes were collected from the MycoCosm database.
- Reference core genes were identified using the BUSCO datasets and they were classified based on their function using the OrthoDB database for each reference set.
- Hidden Markov Models (HMMs) were generated for analysis.
- Duplication thresholds, dN/dS, single copy and ubiquity values are identified for each reference genome.
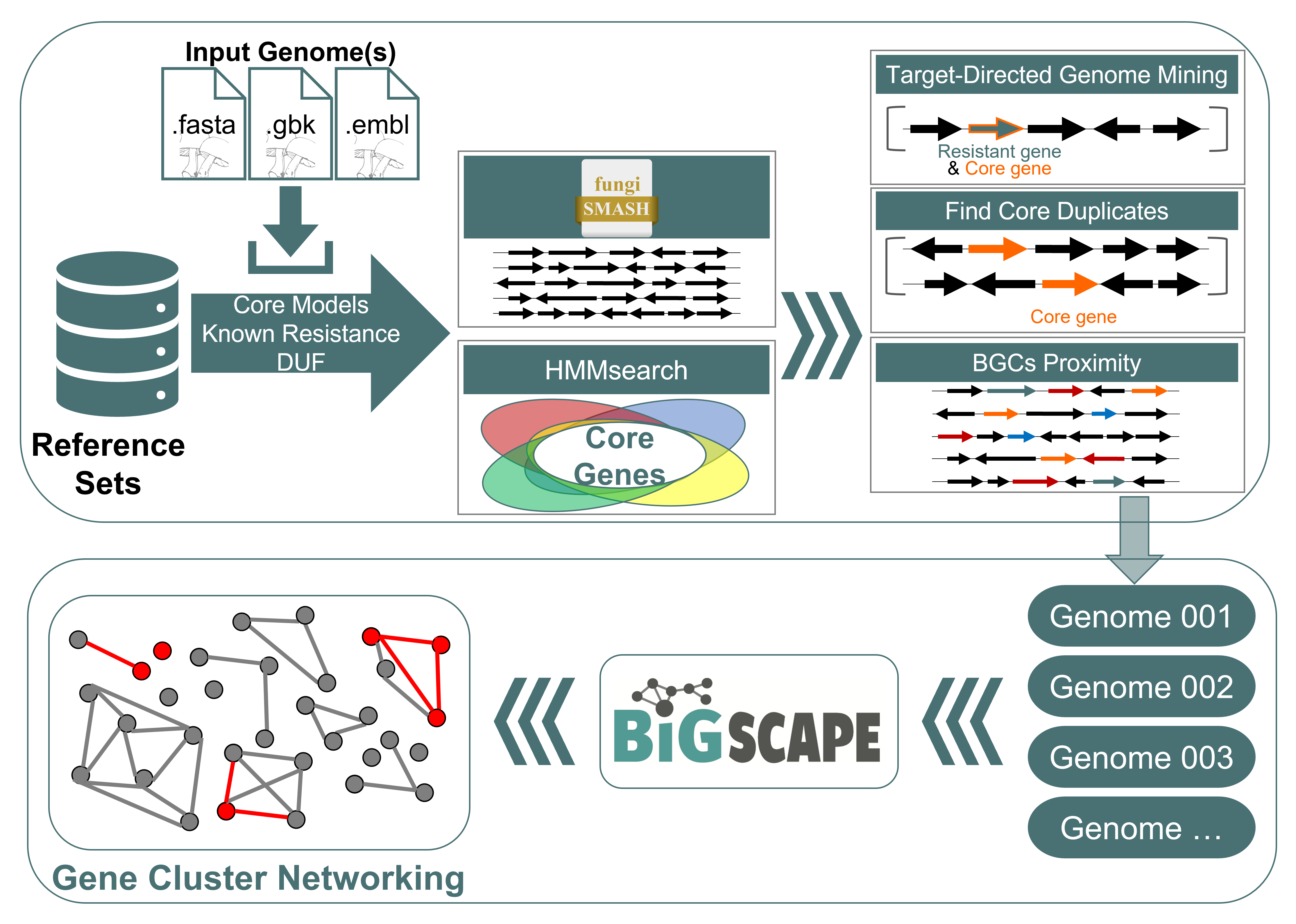
Query genome workflow:
- Biosynthetic Gene Clusters (BGCs) are identified with antiSMASH if this is not already present. (minimal run omitting extra options is performed)
- Known resistance & target models are searched and identified
- Core genome models from reference are used to identify and extract query genes.
- Duplications are marked based on deviation from the sum of the reference median count and standard deviation. Note: Draft genomes with repeat genes due to mis-assembly should be manually confirmed
- To compare the results of multi-genome analysis, the BiG-SCAPE clustering algorithm is applied to all determined BGCs. The BiG-SCAPE algorithm generates sequence similarity networks of BGCs and classifies them into gene cluster families (GCFs).
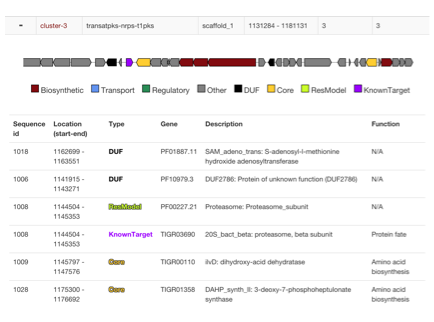
- Biosynthetic Gene Clusters (BGCs) are identified with antiSMASH or read from genbank "cluster" annotations
- Locations of Core genes, DUF, Resistance models and custom models are checked if they are within cluster boundaries
- Additional cluster visualization is presented to identify where hits are present and in what context
If you found FunARTS to be helpful please cite the following publication:
Yılmaz, T. M., Mungan, M. D., Berasategui, A., & Ziemert, N. (2023). FunARTS, the Fungal bioActive compound Resistant Target Seeker, an exploration engine for target-directed genome mining in fungi. Nucleic Acids Research, 10.1093/nar/gkad386
- Thaker, M. N., Wang, W., Spanogiannopoulos, P., Waglechner, N., King, A. M., Medina, R., & Wright, G. D. (2013). Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nature Biotechnology, 31(10), 922–927.
- Tang, X., Li, J., Millán-Aguiñaga, N., Zhang, J. J., O’Neill, E. C., Ugalde, J. A., … Moore, B. S. (2015). Identification of Thiotetronic Acid Antibiotic Biosynthetic Pathways by Target-directed Genome Mining. ACS Chemical Biology, 10(12), 2841–2849.
- Johnston, C. W., Skinnider, M. A., Dejong, C. A., Rees, P. N., Chen, G. M., Walker, C. G., … Magarvey, N. A. (2016). Assembly and clustering of natural antibiotics guides target identification. Nature Chemical Biology, 12(4), 233–239.
- Blin, K., Shaw, S., Kloosterman, A. M., Charlop-Powers, Z., Van Wezel, G. P., Medema, M. H., & Weber, T. (2021). antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Research, 49(W1), W29-W35.
- Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., & Zdobnov, E. M. (2021). BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes, Molecular Biology and Evolution, 38(10), 4647-4654.
- Kriventseva, E. V., Kuznetsov, D., Tegenfeldt, F., Manni, M., Dias, R., Simão, F. A., & Zdobnov, E. M. (2019). OrthoDB v10: sampling the diversity of animal, plant, fungal, protist, bacterial and viral genomes for evolutionary and functional annotations of orthologs, Nucleic Acids Research, 47(D1), D807-D811.
- Mungan,M.D., Alanjary,M., Blin,K., Weber,T., Medema,M.H., & Ziemert,N. (2020). ARTS 2.0: feature updates and expansion of the Antibiotic Resistant Target Seeker for comparative genome mining. Nucleic Acids Research, 48(W1), W546–W552.
- Alanjary,M., Kronmiller,B., Adamek,M., Blin,K., Weber,T., Huson,D., Philmus,B., & Ziemert,N. (2017). The Antibiotic Resistant Target Seeker (ARTS), an exploration engine for antibiotic cluster prioritization and novel drug target discovery, Nucleic Acids Research, 45(W1), W42-W48.